The Unseen Blueprint: How MTHFR Polymorphisms Impact Breast Cancer Risk
October is Breast Cancer Awareness Month. This week I turn 49, which is the age my mother was when she died of breast cancer. This age has been looming large in my future for over 2 decades. When I ran across research talking about breast cancer (as well as other cancers) and MTHFR SNPs, I was intrigued!
As Registered Dietitians and nutrition professionals, we are experts in translating complex metabolic information into actionable wellness plans. Yet, sometimes, the sheer volume of advanced biochemistry we mastered early in our careers, like the intricate details of the one-carbon metabolism pathway, can feel distant – especially when dealing with highly personal health concerns.
Today, we delve into one critical genetic factor relevant to breast cancer susceptibility that ties directly back to core metabolic function: the MTHFR gene (methylenetetrahydrofolate reductase).
MTHFR and the Fundamentals of Methylation
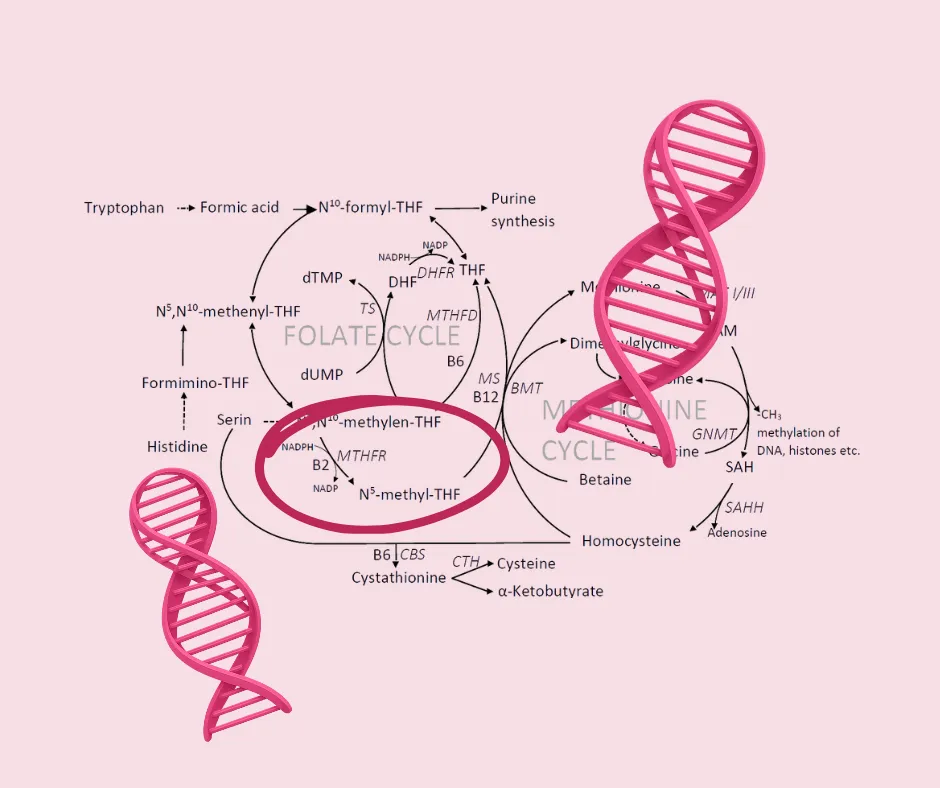
For those who may be a little rusty on the specifics of the one-carbon cycle, MTHFR is an essential enzyme whose function is foundational to numerous processes in the body (Hyman, 2011).
The MTHFR Role in Folate Metabolism: The MTHFR gene encodes the methylenetetrahydrofolate reductase enzyme (Lal et al., 2022; Li et al., 2014; Yan et al., 2016). This enzyme catalyzes the crucial, irreversible conversion of 5,10-methylenetetrahydrofolate (5,10-methylene-THF) to 5-methyltetrahydrofolate (5-methylene-THF), which is the predominant circulating and active form of folate.
The Methylation Cascade: The active folate (5-methylene-THF) provides a methyl group necessary for the remethylation of homocysteine to methionine (Lal et al., 2022; Li et al., 2014). Methionine is the precursor for S-adenosylmethionine (SAMe), which serves as the body’s universal methyl donor. The initiation of this methylation process is so vital that MTHFR is often referred to as the “mother of all genes” because it has a hand in many critical bodily processes (Lynch, 2020; Seeking Health, 2021).
Why Methylation Matters to Cancer Risk: Methylation governs nearly every biochemical process, affecting brain chemistry, energy production, detoxification, antioxidant production, cellular repair, and genetic expression (Hyman, 2011; Lynch, 2020; Seeking Health, 2021). Folate plays an integral role in maintaining DNA stability by regulating DNA biosynthesis, DNA repair, and DNA methylation (Lal et al., 2022; Li et al., 2014). If methylation breaks down or is compromised, it can disrupt DNA repair synthesis and DNA methylation, which may increase cancer risk (Li et al., 2014).
When the Gene is "Born Dirty": MTHFR Polymorphisms
When clients discuss genetic testing, they are often referring to polymorphisms or single nucleotide polymorphisms (SNPs): slight variations in the genetic code. When a genetic variation causes a gene to work at a reduced rate, this is sometimes termed a "born dirty gene," and the MTHFR gene is a key example of this (Lynch, 2020; Seeking Health, 2021).
The two most common MTHFR polymorphisms investigated in relation to health outcomes are 677 C>T (rs1801133) and 1298 A>C (rs1801131) (Lal et al., 2022; Li et al., 2014). These common polymorphisms are associated with reduced enzyme activity (Li et al., 2014):
MTHFR C677T Polymorphism: This variation involves the substitution of a C (cytosine) for a T (thymine) at nucleotide position 677 (Lal et al., 2022; Yan et al., 2016). Individuals homozygous for the T allele (TT genotype) may have MTHFR enzyme activity reduced to approximately 30% compared to the wild-type (CC) (Lal et al., 2022; Li et al., 2014). Heterozygous individuals (CT genotype) typically have about 65% activity (Lal et al., 2022).
MTHFR A1298C Polymorphism: This variation can result in the homozygous CC genotype having about 60% enzyme activity compared to the wild-type (AA) (Li et al., 2014).
A reduced function in MTHFR means the body may produce less active folate (methylfolate) and consequently, fewer methyl groups are available for essential functions like detoxification and DNA repair (MTHFR Support Australia, n.d.).
The Established Link: MTHFR and Breast Cancer Susceptibility
As experts, we know that cancer etiology is complex, involving interactions between low susceptibility genes and environmental factors (Lal et al., 2022; Li et al., 2014). When the association between MTHFR polymorphisms and breast cancer risk was studied, early results were often inconclusive, prompting large-scale meta-analyses to draw a more robust conclusion (Lal et al., 2022; Li et al., 2014).
Here is what large, comprehensive meta-analyses suggest regarding the two primary MTHFR polymorphisms and breast cancer risk:
1. MTHFR 677 C>T Polymorphism
The association between the MTHFR C677T polymorphism and breast cancer risk has been consistently identified in meta-analyses (Yan et al., 2016).
A systematic meta-analysis combining 57 studies concluded that the MTHFR 677 C>T polymorphism may contribute to breast cancer development (Li et al., 2014).
Overall analysis of these studies demonstrated a statistically significant association with breast cancer risk (Li et al., 2014; Yan et al., 2016).
The 677 T allele is specifically identified as a low-penetrance risk factor (a genetic variance that increases risk for a disease but usually does not increase risk as much as other genetic variations) for developing breast cancer (Yan et al., 2016). This association appears particularly strong, and often statistically significant, in Asian populations (Lal et al., 2022; Li et al., 2014).
2. MTHFR 1298 A>C Polymorphism
While the 677 C>T variant shows a clearer association, the findings for the 1298 A>C polymorphism are less consistent across the total population studied:
Overall, the A1298C polymorphism has been found not to be significantly associated with increased risk of breast cancer (Li et al., 2014).
However, subgroup analysis based on ethnicity sometimes yields different results. For example, some studies found that the 1298A allele had a significant effect on breast cancer risk in the Caucasian population (Li et al., 2014).
Integrating Nutrigenomics into Professional Practice
The journey of understanding one’s own health risks, especially those with a strong family history, requires research-informed encouragement. You are not a victim of “bad genes;” thanks to the principles of epigenetics and nutrigenomics, your genes are not your destiny (Lynch, 2020; Seeking Health, 2021).
The knowledge that lifestyle, diet, and environment can highly influence how genes are expressed is profoundly empowering (Lynch, 2020; Seeking Health, 2021). Since the MTHFR gene’s function relates directly to folate status and methylation, factors like diet (minimizing caffeine, alcohol, processed foods, and ensuring sufficient B vitamins and leafy greens), managing stress, and targeted supplementation can support the function of this key pathway (Hyman, 2011; Lynch, 2020; MTHFR Support Australia, n.d.; Seeking Health, 2021).
If you are questioning your career path or feeling overwhelmed, consider that diving deeper into nutrigenomics and genetic interpretation could be a powerful, high-level pivot point that utilizes your existing expertise in biochemistry and metabolism. Resources like genetic testing and educational courses on nutrigenomics are available to enhance your knowledge and guide personalized clinical applications (MTHFR Support Australia, n.d.; Seeking Health, 2021).
Knowledge truly is power, and when applied to your health or the health of your clients, it can be the most powerful medicine.
Works Cited
Hyman, M. (2011, February 9). Maximizing Methylation: The Key to Healthy Aging. Mark Hyman, MD. https://drhyman.com/blogs/content/maximizing-methylation-the-key-to-healthy-aging-2
Lal, H., Sharma, B., Sambyal, V., Guleria, K., Singh, N., Uppal, M., Manjari, M., Sudan, M., Singh, N. R., & Uppal, M. S. (2022). Association of 677C>T polymorphism with breast cancer risk: A case-control study and meta-analysis. Journal of Cancer Research & Therapeutics, 18(6), 1451–1460. https://doi.org/10.4103/jcrt.JCRT_1063_20
Li, K., Li, W., & Dong, X. (2014). Association of 677 C>T (rs1801133) and 1298 A>C (rs1801131) Polymorphisms in the MTHFR Gene and Breast Cancer Susceptibility: A Meta-Analysis Based on 57 Individual Studies. PLoS ONE, 9(6), 1–9. https://doi.org/10.1371/journal.pone.0071290
Lynch, B. (2020). Dirty Genes. Harper One.
MTHFR Support Australia. (n.d.). What is MTHFR. MTHFR Support Australia. Retrieved October 18, 2025, from https://www.mthfrsupport.com.au/what-is-mthfr/
Seeking Health. (2021, November 12). Dirty Genes®: How to Change Your Genetic Destiny. Seeking Health. https://www.seekinghealth.com/blogs/education/dirty-genes?srsltid=AfmBOooHIuGwWHfldYKJ7vjkawHUqtVrIyUGEWJ5hlPXMZ2CfKMMkMKj
Yan, W., Zhang, Y., Zhao, E., & Zhang, S. (2016). Association between the MTHFR C677T Polymorphism and Breast Cancer Risk: A Meta-Analysis of 23 Case–Control Studies. The Breast Journal, 22(5), 593–594. https://doi.org/10.1111/tbj.12639
